
Every cell in the human body contains the same genetic instructions for making and maintaining an individual. What differs across cell types — from liver cells to heart cells to cancer cells — is which parts of a person’s DNA are read.
A University of Texas at Dallas scientist has received a five-year, $1.9 million federal grant to support research aimed at better understanding how access to the DNA in human cells is regulated.
Dr. Sheena D’Arcy, assistant professor of chemistry in the School of Natural Sciences and Mathematics, recently received a Maximizing Investigators’ Research Award for Early Stage Investigators (ESI-MIRA) (grant number R35GM133751) from the National Institute of General Medical Sciences (NIGMS), a component of the National Institutes of Health (NIH).
D’Arcy’s research focuses on the structure and function of the proteins that are involved in gene transcription — the process of “reading” DNA instructions to create the proteins that carry out life functions.
If the DNA in a single cell was stretched out, it would be about 6 feet long. To store DNA in an ordered way and to regulate access to it, it is coiled and wrapped in structures called nucleosomes, which are each made up of several proteins called histones.
When it comes time for the transcription machinery to read or copy genetic information, the nucleosome and its histones must disassemble to allow access and then reassemble — like a thread being unwrapped from a spool and wrapped up again. This complex process is controlled by proteins such as histone chaperones.
“We don’t need to read all of the DNA all of the time, just parts of it,” D’Arcy said.
“Proteins aren’t static; they change shape and move as they interact with other proteins and molecules. We can’t directly see the molecules moving, but we can use this technique to get a better picture of the dynamics. It’s a niche approach, and not many people can do it, which I think contributed to us receiving this competitive grant.”
Dr. Sheena D’Arcy, assistant professor of chemistry in the School of Natural Sciences and Mathematics
Citing an analogy championed by her former mentor, D’Arcy added: “Like a chaperone at a school dance, histone chaperones make sure histones have ‘appropriate’ interactions at the appropriate time.”
Under the new NIH grant, D’Arcy is studying various proteins and protein modifications that influence the assembly and disassembly of the nucleosome. Insights gained could have an impact on combating diseases such as cancer.
“Generally in cancer, the parts of the DNA that are read can change because of changes in the nucleosome structure,” she said. “The proteins that change nucleosome structure are drug targets, so understanding these interactions is useful for designing new drugs.”
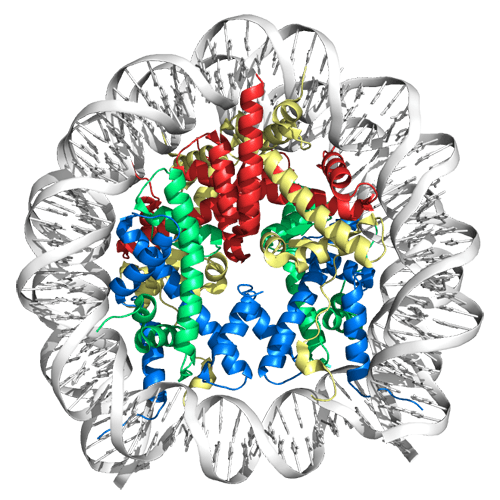
D’Arcy uses an advanced technology called hydrogen/deuterium exchange mass spectroscopy to track over time how nucleosomes change when they interact with other proteins, resulting in a type of “movie” of the process.
“Proteins aren’t static; they change shape and move as they interact with other proteins and molecules. We can’t directly see the molecules moving, but we can use this technique to get a better picture of the dynamics,” D’Arcy said. “It’s a niche approach, and not many people can do it, which I think contributed to us receiving this competitive grant.”
D’Arcy’s grant supported her work in a study published in November in the journal Nature. She and chemistry doctoral student Naifu Zhang demonstrated the power of hydrogen/deuterium exchange mass spectroscopy by observing a histone chaperone called “facilitates chromatin transcription,” or FACT, carrying out its essential role.
“In this research, we have essentially frozen the FACT protein in the middle of it changing the nucleosome, although we aren’t sure whether it is unwrapping or wrapping the nucleosome,” D’Arcy said.
About MIRA Grants
Unlike a traditional research grant, MIRA grants from the NIH offer investigators the flexibility to pursue a broader scientific question as it evolves, rather than being tied to one specific project. The grant program is part of the NIH’s strategy to bring innovation and risk-taking back to basic medical research.
D’Arcy and Zhang became involved in the FACT project when researchers at the University of Colorado Boulder, who led the effort, turned to D’Arcy’s lab for assistance. The Colorado group and researchers at the National Resource for Automated Molecular Microscopy, part of the NIGMS, used a technique called cryo-electron microscopy (cryo-EM) to obtain a high-resolution image of FACT bound to a nucleosome.
“Our colleagues in Colorado reached out to us because of our expertise in studying proteins in solution,” D’Arcy said. “Our role was to use hydrogen/deuterium exchange mass spectroscopy to see if the solution behavior of the complex matched what they’d observed in the structure.
“They had the cryo-EM images, and we had the solution studies. We didn’t share our results until we both had complete data, and then the data were 100% consistent. It was one of those ‘wow’ moments. These new findings about the FACT protein give us a much better picture of FACT’s role in regulating access to DNA. It was also great to collaborate with two other woman-led teams.”